📋 Content
We will not go into the physical principles of image creation in this chapter!
Overview
Biomedical image analysis is a field that focuses on extracting meaningful information from medical images. These images capture various aspects of the human body, ranging from microscopic to macroscopic scales. Here are some key points about biomedical image analysis:
- Types of Biomedical Images:
- Biomedical images are acquired using different imaging modalities, such as CT (computed tomography), MRI (magnetic resonance imaging), ultrasound, and X-rays.
- These images represent physical properties of the body, such as radiodensity (how well tissues absorb X-rays) or tissue structure.
- Clinical Importance:
- Domain experts, such as radiologists, analyze these images for clinical purposes.
- Biomedical images play a crucial role in medical decision-making, aiding in diagnoses, treatment planning, and monitoring patient progress.
- Image Characteristics:
- Biomedical images are often volumetric (3D), capturing structures in three dimensions.
- Some images may have an additional time dimension (4D) or multiple channels (4-5D), as seen in multi-sequence MRIs.
- Unlike natural images (e.g., photographs), biomedical images follow specific acquisition protocols (e.g., patient positioning) to ensure consistency.
- Challenges in Analysis:
- Detecting subtle differences is essential. For instance, identifying small regions indicating abnormalities.
- Researchers and practitioners develop algorithms to automate tasks like segmentation (identifying regions of interest), registration (aligning images), and feature extraction.
In summary, biomedical image analysis bridges the gap between medical imaging technology and clinical practice.
Load images
There are many ways to load a corresponding DICOM image (2D) or later an entire volume (3D), one of which is offered by the imageio package. To warm up, we load a single DICOM image from the scan volume and check out a few of its attributes:
im = imageio.imread('dataset/slice_000.dcm')
# Print image attributes
print('Image type:', type(im))
print('Shape of image array:', im.shape)
Image type: <class 'imageio.core.util.Array'>
Shape of image array: (256, 256)
Metadata
ImageIO reads in data as Image objects. These are standard NumPy arrays with a dictionary of metadata.
Metadata can be quite rich in medical images and can include:
- Patient demographics: name, age, sex, clinical information
- Acquisition information: image shape, sampling rates, data type, modality (such as X-Ray, CT or MRI)
Plot images
The most critical principle of image analysis is: look at your images!
Matplotlib’s imshow() function gives you a simple way to do this. Knowing a few simple arguments will help:
cmapcontrols the color mappings for each value. The “gray” colormap is common, but many others are available.vminandvmaxcontrol the color contrast between values. Changing these can reduce the influence of extreme values.plt.axis('off')removes axis and tick labels from the image.
Load volumes
ImageIO’s volread() function can load multi-dimensional datasets and create 3D volumes from a folder of images. It can also aggregate metadata across these multiple images.
vol = imageio.volread('dataset/')
# Print image shape
print('Shape of image array:', vol.shape)
Reading DICOM (examining files): 1/192 files
Found 1 correct series.
Reading DICOM (loading data): 192/192 (100.0%)
Shape of image array: (192, 256, 256)
Field of view
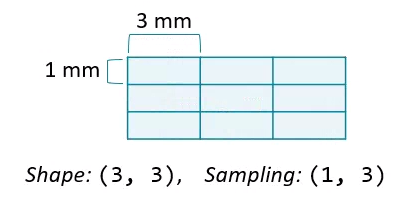
The amount of physical space covered by an image is its field of view, which is calculated from two properties:
- Array shape, the number of data elements on each axis.
- Sampling resolution, the amount of physical space covered by each pixel.
Data Normalization
Data normalization is a crucial step in preparing biomedical image data for analysis. Its purpose is to reduce unwanted variations while preserving relevant information. Here are the most common forms of normalization:
- Intensity Normalization:
- Biomedical images often exhibit variations in intensity due to factors like imaging equipment, patient positioning, and lighting conditions.
- Intensity normalization aims to standardize pixel values across different images.
- Spatial Normalization:
- Biomedical images may have variations in spatial orientation (e.g., subject pose, rotation, and scaling).
- Spatial normalization aligns images to a common reference space. Techniques include:
- Affine transformations: Rotation, translation, and scaling to align images.
- Non-rigid registration: Deformable transformations to account for local variations.
- Contrast Enhancement:
- Enhancing image contrast improves visibility of structures.
- Methods include histogram equalization, adaptive histogram equalization, and gamma correction.
- Tissue-Specific Normalization:
- Some tissues or regions of interest may require specific normalization.
- For instance, normalizing brain images might involve specific brain tissue segmentation and normalization within each tissue type.